
By Dr. Linda Jacobson, Toronto Humane Society/OSMA
In the shelter, we frequently refer to Dr. Scott Weese’s diagnostic advice: “Never do a test without a (good) reason” and “Never do a test without a plan to use the result”. To this one might add: “Never do a test whose results you are unable to interpret”.
This last might seem odd in the context of PCR results – aren’t they just a straightforward positive or negative? Not really! The exquisite sensitivity of PCR and the ubiquity of test panels represent both opportunities and pitfalls in the diagnosis of infectious diseases.
PCR Basics
qPCR tests use a defined number of DNA amplification cycles, typically 40. The cycle threshold value (Ct value) is the number of cycles required to be able to detect the fluorescent signal from the amplified DNA. The more pathogen in the initial sample, the fewer cycles required for detection. The Ct value is therefore inversely proportional to the amount of pathogen in the sample and represents an exponential process. Typically, a large amount of pathogen would yield Ct values of about 15-25, while a small amount of pathogen would yield Ct values of 35-39.99.
PCR can detect as few as 5-10 initial copies of a pathogen, in amounts far too low to be infective and often not reflecting disease caused by that organism. Overdiagnosis is therefore a major potential pitfall. Assuming an otherwise reliable test, negative PCR results from a positive animal will only occur if:
- there was no pathogen in the sample submitted
- the amount of pathogen falls below a diagnostic cut-off, or
- the sample was too small or incorrectly stored.
“False positive” results can result from
- laboratory error
- sample contamination, or
- if the pathogen is indeed present, but the disease is not
True false positive results are not possible for the pathogen, because PCR primers are highly specific, but a “false positive” diagnosis of the disease is certainly possible. It can generally be assumed that the higher the Ct value, the less likely an animal is to be contagious. If there is uncertainty about the results, Ct values should be requested from the laboratory.
Lessons from COVID-19
qPCR can detect RNA or DNA from non-viable pathogens. In early work on SARS-CoV-2, viral RNA was detected on a high proportion of surfaces in the vicinity of COVID-19 patients, but zero samples (of a total of 97) contained infectious titres of virus on tissue culture.
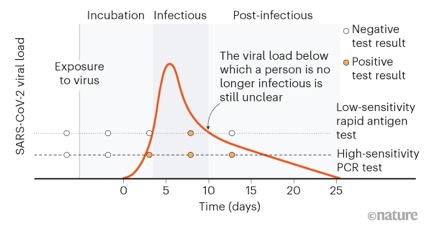
COVID-19 has substantially improved our understanding of the relationship between rapid antigen tests and qPCR. This relationship is very similar for parvo and panleukopenia. Positive rapid test results are reliable, but negative results may not be. This is because the antigen test requires high viral loads for a positive result. In both dogs and cats, parvoviral counts peak within several days of infection (typically 3-7 days). Antigen tests are positive in the early, most acute stages of infection, while qPCR typically remains positive for longer. The rapid test represents a “tip of the iceberg” phenomenon, and is positive at the peak of virus load and infectivity. The rapid test is recommended as the primary test for diagnosing parvo and panlekopenia infection, with qPCR only required under specific circumstances.
Canine parvo and feline panleukopenia (aka feline parvovirus)
Low-level, clinically insignificant, shedding of parvoviruses is quite common, particularly after modified live virus vaccination. Virus can also be detected for weeks post-infection, long after virus isolation (demonstrating viability) is negative. To avoid misdiagnosis, so-called technical cut-off Ct values are set, below which results are reported as positive, and above which they are reported as negative. The cut-off for the IDEXX canine and feline parvo qPCR is Ct ≤ 26 (~1.6 x 106 virus particles per gram of feces). Virus levels close to the cut-off may still be clinically significant. For this reason, Ct values should ideally be reported for all results, and should be requested if they are not provided and results are difficult to interpret.
Positive qPCR results may occur in the first 3-7 days after an initial modified live virus vaccination. This may pose a diagnostic dilemma. A false positive diagosis of parvo or panleukopenia could lead to unnecessary isolation or even euthanasia. Vaccine positives are much less likely for rapid antigen tests (such as the IDEXX SNAP test), and these are recommended for first-line testing. qPCR should be reserved for animals that are strongly suspected to be infected but have a negative rapid test, or for specific surveillance purposes in high-risk groups.
Dermatophytosis/ringworm
qPCR is highly sensitive for diagnosis of Microsporum canis infection in shelter cats, with the enormous advantage of allowing rapid diagnosis, and rapid rule-out for the many suspects that are ultimately negative. Results are available in 2-3 days, compared with 2 weeks for fungal culture.
Cats with no skin lesions and a positive PCR test are likely to be so-called “fomite carriers”, having coat contamination from the environment. This is common in settings with low-level endemic infection. Fomite carriers are distinguished from true positives by the lack of skin lesions and rapid conversion to negative culture status. While semi-quantitative culture is used to distinguish fomite carriers from true positives, no reliable qPCR cut-off has been established. However, a high Ct value would be consistent with carrier status.
In cats being treated for Microsporum canis infection, qPCR remained positive long after fungal cultures were negative. Ct values were generally unhelpful in predicting when cultures would be negative, other than that values ≥ 35.7 were associated with negative cultures. Prolonged positive qPCR results could be interpreted to mean that a cat is still infected and contagious. This could lead to unnecessarily prolonged treatment and isolation, extra clinical interventions and additional outbreak measures. For these reasons, weekly fungal culture is recommended in shelters after the initial qPCR.
Pitfalls of PCR panels
qPCR tests are increasingly offered as multiplex test panels, which simultaneously test for a number of pathogens. The advantage of this is time, cost, and avoidance of tunnel vision and confirmation bias. However, interpretation of test panels requires an understanding of the likelihood of asymptomatic shedding, the clinical significance of a positive test and the need for intervention. In some contexts, particularly in shelter settings, results may be uninterpretable. For example, feline upper respiratory pathogens were commonly detected in shelter cats without clinical signs of URI, and enteropathogens were frequently identified in feces from shelter dogs and cats both with or without diarrhea. Incorrect interpretation can lead to unnecessary interventions, additional testing, and barriers to adoption in a shelter setting. Clinical guidelines, providing clear case definitions and other guidance, would be of substantial benefit. Outcomes-based studies are also needed, as the benefits of assay panels in human medicine are not clearly established.
Back to Basics
Not all pathogens cause disease, not all identified pathogens are the cause of disease in a given animal, and not all pathogens need to be treated. It is extremely important to be able to assess which is which, in the context of qPCR. This leads us back to first principles: assess the context, history, clinical presentation and consequences of action (or inaction), as well as the laboratory result. That result, however authoritative it may seem, is only one piece of information among many.
More information
- Kralik P, Ricchi M. A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol 2017; 8: 1–9.
- Freisl M, Speck S, Truyen U, et al. Faecal shedding of canine parvovirus after modified-live vaccination in healthy adult dogs. Vet J 2017; 219: 15–21.
- Jacobson L, Janke K, Ha K, et al. Feline panleukopenia virus DNA shedding following modified live virus vaccination in a shelter setting. Vet J 2022; 279: 105783.
- Janke KJ, Jacobson LS, Giacinti JA, et al. Fecal viral DNA shedding following clinical panleukopenia infection in shelter kittens: a prospective, observational study. J Feline Med Surg 2021; 24: 337–343.
- Jacobson LS, Janke KJ, Giacinti J, et al. Diagnostic testing for feline panleukopenia in a shelter setting: a prospective, observational study. J Feline Med Surg 2021; 23: 1192–1199.
- Jacobson LS, Mcintyre L, Mykusz J. Comparison of real-time PCR with fungal culture for the diagnosis of Microsporum canis dermatophytosis in shelter cats : a field study. J Feline Med Surg 2018; 20:103-107
- Ben-Shmuel A, Brosh-Nissimov T, Glinert I, et al. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clin Microbiol Infect 2020; 26: 1658–1662.
- Jacobson LS, McIntyre L, Mykusz J. Assessment of real-time PCR cycle threshold values in Microsporum canis culture-positive and culture-negative cats in an animal shelter: a field study. J Feline Med Surg 2018; 20: 108–113.
Positivity for rapid antigen test compared with PCR test, for SARS-CoV-2
The CASCMA Blog is proudly sponsored by Ogena Solutions

